Imports: Make sure these are installed in your conda env, and that the conda env is active in your notebook
numpy
matplotlib
scipy
jupyter
For interactive plots:
ipykernel
A note on cloud-hosted notebooks. If you are running a notebook on a cloud provider, such as Google Colab or CodeOcean, remember to save your work frequently. Cloud notebooks will occasionally restart after a fixed duration, crash, or prolonged inactivity, requiring you to re-run code.
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
%load_ext autoreload
%autoreload 2The Abelian sandpile¶
Background¶
We are going to implement the celebrated Bak-Tang-Wiesenfeld model, also known as the “Abelian sandpile.” This cellular automaton describes a lattice in which sand grains are continuously dropped onto random locations at a fixed rate, resulting in the formation of random sandpiles. When the sandpiles grow too high, they topple, resulting in avalanches that distribute grains to all of their neighbors.
If we denote the number of grains at a site as , a single “topple” event of the BTW model has the following update rule, which triggers only when .
A topple event thus consists of a single site decreasing in height by four grains, and then distributing these grains to each of its north, south, east, and west neighbors---but not its diagonal neighbors.
After a single topple event, the entire lattice is checked again to see if any other sites now have , in which case those sites are toppled, too. If a site located at the boundary of the domain topples, then any grains that would go out of bounds are assumed to be permanently lost from the system. After all sites reach a state where , the avalanche has concluded. We then add a sand grain to a random new site, and calculate any new resulting avalanches.
This idealized system has several interesting properties: the continuous addition of grains represents a slow-timescale driving process, which effectively injects energy into the system. The avalances represent fast-timescale response dynamics, and the grains that fall off the edges represent dissipation---they prevent avalanches from continuing forever undriven. Despite its seeming simplicity, the BTK model represents perhaps the earliest widely-studied toy model of “self-organized criticality,” a well-known hypothesis in nonequilibrium statistical physics that driven, dissipative systems tend to tune themselves into maximally-critical states (there are always sandpiles on the verge of toppling). The sandpile has been used as a thought experiment describing an incredible array of diverse systems, including: starts and stops in dragging friction, earthquakes, timings between geyser eruptions, timings of pulsar glitches, fluctuations in an ultracold atomic gas, neuronal activity patterns in the brain, flux pinning, and many other areas.
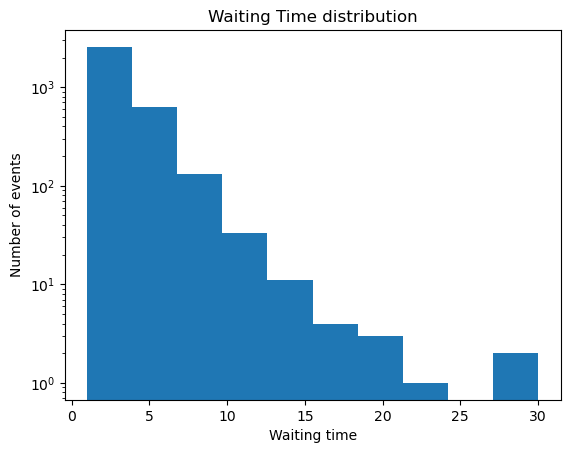
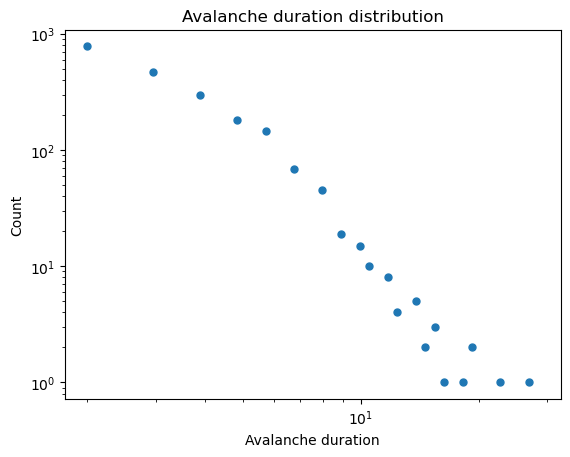
A simple signature of self-organized-criticality observed in the sandpile model, which also represents an easy readout for experimental data, is the appearance of noise in the system’s power spectrum, where denotes frequency. This distribution implies that avalanche durations have a skewed distribution, where larger events happen less frequently. For an experimental system, this results in “crackling” or brown noise in the detector. In the BTW model, the authors note that a distribution of rare events in the frequency corresponds to a distribution of waiting times between rare events.
Here we will implement the BTW model, and then test for the appearance of noise.
To Do:¶
Please complete the following tasks and answer the included questions. You can edit a Markdown cell in Jupyter by double-clicking on it. To return the cell to its formatted form, press [Shift]+[Enter].
Implement the Abelian Sandpile model and simulate its dynamics. I’ve included a template of the iterative solution below, although feel free to re-factor if you would prefer to implement the problem a different way. There are at least three different ways to implement the Abelian sandpile:
The iterative solution involves adding a grain and then repeatedly checking the lattice for piles to topple
The recursive depth-first-search solution adds a grain and then traces the avalanche that results from each grain toppled by the initial addition.
The breadth-first-search solution adds a grain and then simultaneously tracks the four potential avalanches that result from a single grain falling.
Your Answer: Fill in the code below.Using the code included below, show that avalanche durations exhibit a distribution, where is some constant.
Your Answer: Fill in the code below.If a single image of the sandpile has size (the total number of lattice sites), what do you expect to be the worst-case scaling of the runtime of your algorithm? What about the expected scaling of memory usage?
Your Answer:You may have noticed that the waiting time and avalanche size distribution exhibit anomalous scaling in their tails, as visible as a small second peak near the extreme end of the distribution. What causes this effect?
Your Answer:(Optional, Hard Problem). Try vectorizing your sandpile implementation, in order to reduce the number of “for” loops used in your implementation. How does vectorization affect the runtime?
Your Answer: Fill in the code below.class AbelianSandpile:
"""
An Abelian sandpile model simulation. The sandpile is initialized with a random
number of grains at each lattice site. Then, a single grain is dropped at a random
location. The sandpile is then allowed to evolve until it is stable. This process
is repeated n_step times.
A single step of the simulation consists of two stages: a random sand grain is
dropped onto the lattice at a random location. Then, a set of avalanches occurs
causing sandgrains to get redistributed to their neighboring locations.
Parameters:
n (int): The size of the grid
grid (np.ndarray): The grid of the sandpile
history (list): A list of the sandpile grids at each timestep
"""
def __init__(self, n=100, random_state=None):
self.n = n
np.random.seed(random_state) # Set the random seed
self.grid = np.random.choice([0, 1, 2, 3], size=(n, n))
self.history =[self.grid.copy()] # Why did we need to copy the grid?
def step(self):
"""
Perform a single step of the sandpile model. Step corresponds a single sandgrain
addition and the consequent toppling it causes.
Returns: None
"""
########## YOUR CODE HERE ##########
raise NotImplementedError
#
#
# My solution starts by dropping a grain, and then solving for all topple events
# until the sandpile is stable. Watch your boundary conditions carefully.
# We will use absorbing boundary conditions: excess sand grains fall off the edges
# of the grid.
#
#
########## YOUR CODE HERE ##########
# we use this decorator for class methods that don't require any of the attributes
# stored in self. Notice how we don't pass self to the method
@staticmethod
def check_difference(grid1, grid2):
"""Check the total number of different sites between two grids"""
return np.sum(grid1 != grid2)
def simulate(self, n_step):
"""
Simulate the sandpile model for n_step steps.
"""
########## YOUR CODE HERE ##########
raise NotImplementedError
#
#
# YOUR CODE HERE. You should use the step method you wrote above.
#
#
########## YOUR CODE HERE ##########
Test and use your code¶
You don’t need to write any code below, these cells are just to confirm that everything is working and to play with your sandpile implementation
# Run sandpile simulation
model = AbelianSandpile(n=100, random_state=0)
plt.figure()
plt.imshow(model.grid, cmap='gray')
plt.title("Initial State")
model.simulate(10000)
plt.figure()
plt.imshow(model.grid, cmap='gray')
plt.title("Final state")
# Compute the pairwise difference between all observed snapshots. This command uses list
# comprehension, a zip generator, and argument unpacking in order to perform this task
# concisely.
all_events = [model.check_difference(*states) for states in zip(model.history[:-1], model.history[1:])]
# remove transients before the self-organized critical state is reached
all_events = all_events[1000:]
# index each timestep by timepoint
all_events = list(enumerate(all_events))
# remove cases where an avalanche did not occur
all_avalanches = [x for x in all_events if x[1] > 1]
all_avalanche_times = [item[0] for item in all_avalanches]
all_avalanche_sizes = [item[1] for item in all_avalanches]
all_avalanche_durations = [event1 - event0 for event0, event1 in zip(all_avalanche_times[:-1], all_avalanche_times[1:])]
## Waiting time distribution
waiting_times = np.diff(np.array(all_avalanche_times))
plt.figure()
plt.semilogy()
plt.hist(waiting_times)
plt.title('Waiting Time distribution')
plt.xlabel('Waiting time')
plt.ylabel('Number of events')
## Duration distribution
log_bins = np.logspace(np.log10(2), np.log10(np.max(all_avalanche_durations)), 50) # logarithmic bins for histogram
vals, bins = np.histogram(all_avalanche_durations, bins=log_bins)
plt.figure()
plt.loglog(bins[:-1], vals, '.', markersize=10)
plt.title('Avalanche duration distribution')
plt.xlabel('Avalanche duration')
plt.ylabel('Count')
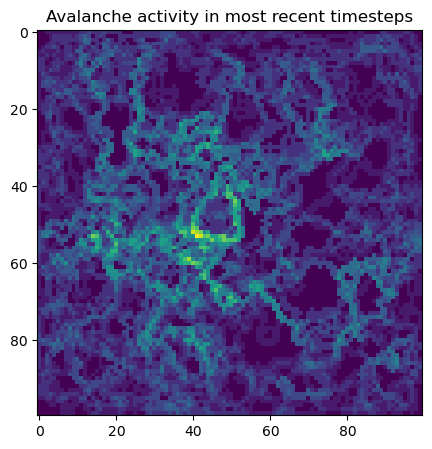
## Visualize activity of the avalanches
# Make an array storing all pairwise differences between the lattice at successive
# timepoints
all_diffs = np.abs(np.diff(np.array(model.history), axis=0))
all_diffs[all_diffs > 0] = 1
all_diffs = all_diffs[np.sum(all_diffs, axis=(1, 2)) > 1] # Filter to only keep big events
most_recent_events = np.sum(all_diffs[-100:], axis=0)
plt.figure(figsize=(5, 5))
plt.imshow(most_recent_events)
plt.title("Avalanche activity in most recent timesteps")Running with Instructor Solutions. If you meant to run your own code, do not import from solutions

Additional information¶
The BTK model is called the “Abelian” sandpile because the final stable configuration is invariant to the order of toppling events. For example, instead of toppling the pile to completion after each addition, we could add several grains at once, and then perform a series of topples selecting unstable sites in any order until a stable configuration is achieved. The final sandpile would be identical to the one we would get if we simulated the pile through full additions sequentially. The Abelian property also means that we have a variety of choices of algorithms for implementing the toppling process resulting from the addition of a single grain; the sandpile will return the same solution regardless of whether a breadth-first search, depth-first search, or iterative solution is used. Importantly, each of these solution methods is, in principle, simulating a different physical process and chain of events, but the particular Abelian property of this model means that the final solution is identical.
Another member of our department, Michael Marder, has a paper from 1992 in which he and his student Steve Kramer propose a cellular automaton model of river formation.. The update rules and structure of implementing this simulation have many simularities to the BTK model.
Among cellular automata, the BTW model is not only unique for its asynchronous nature, but also because it has a “derivative” ruleset in contrast to the “integral”/totalistic ruleset seen in better-known systems like the Game of Life: instead of updating a cell based on the sum of all its neighbors, we update all neighbors based on the state of a cell. A similar distinction arises in other types of dynamical systems; for example, when numerically integrating partial differential equations, finite difference methods update each lattice point based on their differences from neighboring points, approximating spatial derivatives. However, for many classes of partial differential equations, finite element methods instead update each lattice point based on the weighted average of all its neighbors, approximating spatial integrals.
Extra code for animations¶
No need to run this, but this code allows you to visualize the sandpile as it evolves over time.
from matplotlib.animation import FuncAnimation
from IPython.display import HTML
activity_sliding2 = all_diffs[-500:]
vmin = np.percentile(activity_sliding2, 1)
# vmin = 0
vmax = np.percentile(activity_sliding2, 99.8)
# Assuming frames is a numpy array with shape (num_frames, height, width)
frames = np.array(activity_sliding2).copy()
fig = plt.figure(figsize=(6, 6))
img = plt.imshow(frames[0], vmin=vmin, vmax=vmax);
plt.xticks([]); plt.yticks([])
# tight margins
plt.margins(0,0)
plt.gca().xaxis.set_major_locator(plt.NullLocator())
def update(frame):
img.set_array(frame)
ani = FuncAnimation(fig, update, frames=frames, interval=50)
HTML(ani.to_jshtml())all_diffs = np.abs(np.diff(np.array(model.history), axis=0))
# all_diffs = all_diffs[np.sum(all_diffs, axis=(1, 2)) > 1] # Filter to only keep big events
# Use a trick to calculate the sliding cumulative sum
activity_cumulative = np.cumsum(all_diffs, axis=0)
# activity_sliding = activity_cumulative[50:] - activity_cumulative[:-50]
activity_sliding = all_diffs
plt.figure(figsize=(5, 5))
plt.imshow(activity_sliding[-1])# This code saves the sliding cumulative sum as a movie. No need to run this cell
activity_sliding2 = activity_sliding[-500:]
vmin = np.percentile(activity_sliding2, 1)
# vmin = 0
vmax = np.percentile(activity_sliding2, 99.8)
for i in range(len(activity_sliding2) - 1):
out_path = "private_dump/sandpile/frame" + str(i).zfill(4) + ".png"
plt.figure()
plt.imshow(activity_sliding2[i], vmin=vmin, vmax=vmax)
ax = plt.gca()
ax.set_axis_off()
ax.xaxis.set_major_locator(plt.NullLocator())
ax.yaxis.set_major_locator(plt.NullLocator())
ax.set_aspect(1, adjustable='box')
plt.savefig(out_path, bbox_inches='tight', pad_inches=0.0, dpi=300)
plt.close()
Percolation¶
The BTK cellular automaton represents a great example of a simple, toy computational model that exhibits non-trivial properties found in other, much more elaborate problems. It can be considered a dynamical universality class, meaning that many problems map onto it, independently of their microscopic details.
Here, we will consider a well-known example of a universality class describing a nonequilibrium phase transition: the directed percolation problem. This problem is usually attributed to Broadbent & Hammersley in 1957.
The basic idea is deceptively simple: given a D-dimensional lattice (for our purposes, a 2D grid), we randomly mark sites as “blocked” with probability . The remaining sites are “open” for water to flow. For a given value of , what is the probability that water poured into the top of the lattice will percolate to the bottom row through a chain of connected open sites? This problem is directed percolation because it has a preferred direction (water flows from top to bottom). We will assume two open sites are connected if one is north, south, east, or west of the other---but not diagonal. This corresponds to von Neumann neighborhood rules.
A percolation simulation should take a binary array, and return True if it percolates, and False if it does not. There are many ways to implement a percolation algorithm, most of which involve simulating the addition of water to the grid.
Iterative Solution. One option would be start with the top row, mark all open sites as “filled,” and then pass to the next row and search for open sites connected to filled sites. Some addtional care is required, however, because water can pass through channels within a row until it reaches sites that are not directly below filled sites. One workaround would be to perform multiple iterative passes until the filled lattice stops changing. Another edge case is the case where water needs to pass through an uphill channel in order to make it to the bottom---this can be solved by passing over the lattice first from top to bottom, and then vice versa.
Depth-first search (DFS). Another simulation option would be a depth-first-search, where we start from each site on the top row and we search for any chain of North/Sout/East/West hops that leads to the other side of the lattice, marking all visited sites as filled with water. This last methods demonstrates the conceptual simularity between directed percolation and solving a maze puzzle; the only difference that the “blocked” sites in a maze are non-random.
Here we are going to implemented a directed percolation model, and then perform experiments with it in order to determine how the percolation probability depends on the fraction of blocked sites .
To Do¶
To Do¶
Please complete the following tasks and answer the included questions. You can edit a Markdown cell in Jupyter by double-clicking on it. To return the cell to its formatted form, press [Shift]+[Enter].
Implement a two dimensional directed percolation model in Python. Below, there is a code outline for the iterative solution. There are at least two ways to solve this problem.
The iterative solution simulates water pouring into the top row, and then iterate over rows and sites. With this solution, there is some difficulty regarding how to handle the case where water can flow through a channel from right to left, even though we normally iterate from left to right. This can be solved by passing over each row twice. However, there is also a the case where water needs to pass uphill through a channel before it can proceed downhill. This can be solved by passing over the lattice from top to bottom, and then vice versa.
The recursive solution traces all possible paths from the top row to the bottom row, similar to solving a labyrinth. To use this solution, you will need to add an additional private method
_flow_recursive(i, j)that contains the recursive logic.
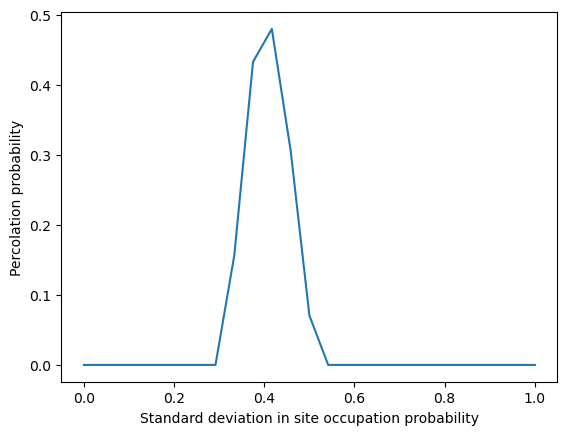
Your Answer: Fill in the code below.Perform replicate simulations using the code provided below, and create a plot showing how the probability of percolation changes as a function of (the blockage probability). Beyond seeing the percolation to clogging transition, you’ll notice that the variance in the outcome of your simulations behaves unexpectedly. What is going on here? (Hint: If you’ve studied the Ising model, you’ve seen something like this before)
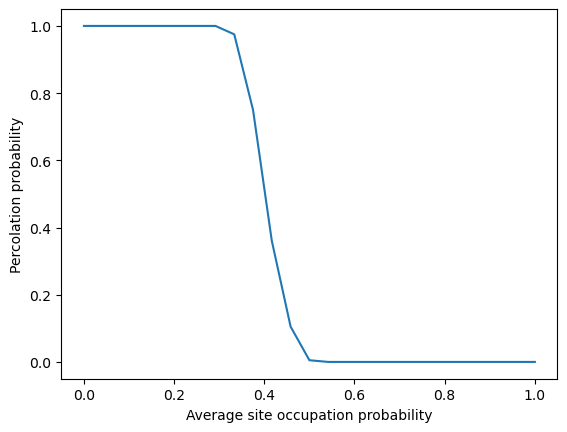
Your Answer: Theoretically, does the transition point seen in our empirical results align with your intuition? Why does its value differ from ? (Hint: think about sites versus connections between sites).
Your Answer: How does the memory usage and runtime of your percolation model implement scale with the lattice size? You can answer this empirically or theoretically.
Your Answer: One way to sweep the control parameter would be to start in a limit where most of the sites are blocked, and then gradually open up individual sites one at a time until the lattice percolates. A video of a simulation where the lattice is gradually opened can be seen here. How do the different events in this gradually unblocked percolation simulation relate to the two timescales we saw in the sandpile problem?
Your Answer: class PercolationSimulation:
"""
A simulation of a 2D directed percolation problem. Given a 2D lattice, blocked sites
are denoted by 0s, and open sites are denoted by 1s. During a simulation, water is
poured into the top of the grid, and allowed to percolate to the bottom. If water
fills a lattice site, it is marked with a 2 in the grid. Water only reaches a site
if it reaches an open site directly above, or to the immediate left or right
of an open site.
I've included the API for my solution below. You can use this as a starting point,
or you can re-factor the code to your own style. Your final solution must have a
method called percolate that creates a random lattice and runs a percolation
simulation and
1. returns True if the system percolates
2. stores the original lattice in self.grid
3. stores the water filled lattice in self.grid_filled
+ For simplicity, use the first dimension of the array as the percolation direction
+ For boundary conditions, assume that any site out of bounds is a 0 (blocked)
+ You should use numpy for this problem, although it is possible to use lists
Attributes:
grid (np.array): the original lattice of blocked (0) and open (1) sites
grid_filled (np.array): the lattice after water has been poured in
n (int): number of rows and columns in the lattice
p (float): probability of a site being blocked in the randomly-sampled lattice
random_state (int): random seed for the random number generator
random_state (int): random seed for numpy's random number generator. Used to
ensure reproducibility across random simulations. The default value of None
will use the current state of the random number generator without resetting
it.
"""
def __init__(self, n=100, p=0.5, grid=None, random_state=None):
"""
Initialize a PercolationSimulation object.
Args:
n (int): number of rows and columns in the lattice
p (float): probability of a site being blocked in the randomly-sampled lattice
random_state (int): random seed for numpy's random number generator. Used to
ensure reproducibility across random simulations. The default value of None
will use the current state of the random number generator without resetting
it.
"""
self.random_state = random_state # the random seed
# Initialize a random grid if one is not provided. Otherwise, use the provided
# grid.
if grid is None:
self.n = n
self.p = p
self.grid = np.zeros((n, n))
self._initialize_grid()
else:
assert len(np.unique(np.ravel(grid))) <= 2, "Grid must only contain 0s and 1s"
self.grid = grid.astype(int)
# override numbers if grid is provided
self.n = grid.shape[0]
self.p = 1 - np.mean(grid)
# The filled grid used in the percolation calculation. Initialize to the original
# grid. We technically don't need to copy the original grid if we want to save
# memory, but it makes the code easier to debug if this is a separate variable
# from self.grid.
self.grid_filled = np.copy(self.grid)
def _initialize_grid(self):
"""
Sample a random lattice for the percolation simulation. This method should
write new values to the self.grid and self.grid_filled attributes. Make sure
to set the random seed inside this method.
This is a helper function for the percolation algorithm, and so we denote it
with an underscore in order to indicate that it is not a public method (it is
used internally by the class, but end users should not call it). In other
languages like Java, private methods are not accessible outside the class, but
in Python, they are accessible but external usage is discouraged by convention.
Private methods are useful for functions that are necessary to support the
public methods (here, our percolate() method), but which we expect we might need
to alter in the future. If we released our code as a library, others might
build software that accesses percolate(), and so we should not alter the
input/outputs because it's a public method
"""
###############################################################################
#
#
####### YOUR CODE HERE #######
raise NotImplementedError("Implement this method")
# Hint: my solution is 3 lines of code in numpy
#
#
###############################################################################
def _flow_recursive(self, i, j):
"""
Only used if we opt for a recursive solution.
The recursive portion of the flow simulation. Notice how grid and grid_filled
are used to keep track of the global state, even as our recursive calls nest
deeper and deeper
"""
####### YOUR CODE HERE #######################################################
raise NotImplementedError("Implement this method")
#
#
# Remember to check the von Neumann neighborhood of the current site. There should
# be 4 recursive calls in total, and 4 base cases
#
#
###############################################################################s
def _poll_neighbors(self, i, j):
"""
Check whether there is a filled site adjacent to a site at coordinates i, j in
self.grid_filled. Respects boundary conditions.
"""
####### YOUR CODE HERE #######################################################
raise NotImplementedError("Implement this method")
#
#
# Hint: my solution is 4 lines of code in numpy, but you may get different
# results depending on how you enforce the boundary conditions in your solution.
# Not needed for the recursive solution
#
#
###############################################################################
def _flow(self):
"""
Run a percolation simulation using recursion
This method writes to the grid and grid_filled attributes, but it does not
return anything. In other languages like Java or C, this method would return
void
"""
###############################################################################
raise NotImplementedError("Implement this method")
####### YOUR CODE HERE #######
# Hintsmy non-recursive solution contains one row-wise for loop, which contains
# several loops over individual lattice sites. You might need to visit each lattice
# site more than once per row. In my implementation, split the logic of checking
# the von neumann neighborhood into a separate method _poll_neighbors, which
# returns a boolean indicating whether a neighbor is filled
#
# My recursive solution calls a second function, _flow_recursive, which takes
# two lattice indices as arguments
###############################################################################
def percolate(self):
"""
Initialize a random lattice and then run a percolation simulation. Report results
"""
###############################################################################
####### YOUR CODE HERE #######
raise NotImplementedError("You must implement this method")
# Hint: my solution is 3 lines of code, and it just calls other methods in the
# class, which do the heavy lifting
###############################################################################
Test and use your code¶
You don’t need to write any new code below, these cells are just to confirm that everything is working and to play with the your percolation implementation
from matplotlib.colors import LinearSegmentedColormap
def plot_percolation(mat):
"""
Plots a percolation matrix, where 0 indicates a blocked site, 1 indicates an empty
site, and 2 indicates a filled site
"""
cvals = [0, 1, 2]
colors = [(0, 0, 0), (0.4, 0.4, 0.4), (0.372549, 0.596078, 1)]
norm = plt.Normalize(min(cvals), max(cvals))
tuples = list(zip(map(norm,cvals), colors))
cmap = LinearSegmentedColormap.from_list("", tuples)
plt.imshow(mat, cmap=cmap, vmin=0, vmax=2)
model = PercolationSimulation(n=20, random_state=0, p=0.1)
print(model.percolate())
plt.figure()
plot_percolation(model.grid_filled)
model = PercolationSimulation(n=20, random_state=0, p=0.4)
print(model.percolate())
plt.figure()
plot_percolation(model.grid_filled)
model = PercolationSimulation(n=20, random_state=0, p=0.9)
print(model.percolate())
plt.figure()
plot_percolation(model.grid_filled)Running with Instructor Solutions. If you meant to run your own code, do not import from solutions
True
Running with Instructor Solutions. If you meant to run your own code, do not import from solutions
True
Running with Instructor Solutions. If you meant to run your own code, do not import from solutions
False
Run replicate simulations across replicates with different bond occupation probabilities
The percolation probability represents an effective order parameter for this system, and so we will attempt to calculate the percolation probability by performing many replicate simulations at different values of the control parameter .
pvals = np.linspace(0, 1, 25) # control parameter for percolation phase transition
n_reps = 200 # number of times to repeat the simulation for each p value
all_percolations = list()
for p in pvals:
print("Running replicate simulations for p = {}".format(p), flush=True)
all_replicates = list()
for i in range(n_reps):
# Initialize the model
model = PercolationSimulation(30, p=p)
all_replicates.append(model.percolate())
all_percolations.append(all_replicates)
plt.figure()
plt.plot(pvals, np.mean(np.array(all_percolations), axis=1))
plt.xlabel('Average site occupation probability')
plt.ylabel('Percolation probability')
plt.figure()
plt.plot(pvals, np.std(np.array(all_percolations), axis=1))
plt.xlabel('Standard deviation in site occupation probability')
plt.ylabel('Percolation probability')
plt.show()
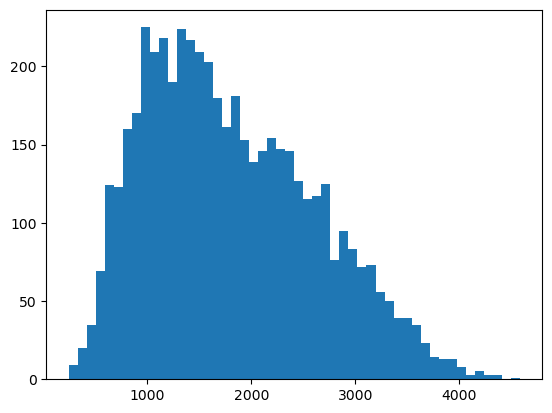
## Just from curiousity, plot the distribution of cluster sizes at the percolation threshold
## why does it appear to be bimodal?
all_cluster_sizes = list()
p_c = 0.407259
n_reps = 5000
for i in range(n_reps):
model = PercolationSimulation(100, p=p_c)
model.percolate()
cluster_size = np.sum(model.grid_filled == 2)
all_cluster_sizes.append(cluster_size)
if i % 500 == 0:
print("Finished simulation {}".format(i), flush=True)
all_cluster_sizes = np.array(all_cluster_sizes)
plt.figure()
plt.hist(all_cluster_sizes, 50);
Running replicate simulations for p = 0.0
Running replicate simulations for p = 0.041666666666666664
Running replicate simulations for p = 0.08333333333333333
Running replicate simulations for p = 0.125
Running replicate simulations for p = 0.16666666666666666
Running replicate simulations for p = 0.20833333333333331
Running replicate simulations for p = 0.25
Running replicate simulations for p = 0.29166666666666663
Running replicate simulations for p = 0.3333333333333333
Running replicate simulations for p = 0.375
Running replicate simulations for p = 0.41666666666666663
Running replicate simulations for p = 0.4583333333333333
Running replicate simulations for p = 0.5
Running replicate simulations for p = 0.5416666666666666
Running replicate simulations for p = 0.5833333333333333
Running replicate simulations for p = 0.625
Running replicate simulations for p = 0.6666666666666666
Running replicate simulations for p = 0.7083333333333333
Running replicate simulations for p = 0.75
Running replicate simulations for p = 0.7916666666666666
Running replicate simulations for p = 0.8333333333333333
Running replicate simulations for p = 0.875
Running replicate simulations for p = 0.9166666666666666
Running replicate simulations for p = 0.9583333333333333
Running replicate simulations for p = 1.0
Finished simulation 0
Finished simulation 500
Finished simulation 1000
Finished simulation 1500
Finished simulation 2000
Finished simulation 2500
Finished simulation 3000
Finished simulation 3500
Finished simulation 4000
Finished simulation 4500
Additional information¶
Directed percolation maps onto all sorts of interesting problems in physics and graph theory. There are very direct applications of percolation to flow in porous media, as well as electrical conductance and light propagation in disordered materials. A great example of the broader relevance of the directed percolation universality class is recent work showing similar scaling exponents describing both ecological collapse and pipe turbulence, which arise due to both systems belonging to the Directed Percolation universality class. Another recent paper uses percolation theory to understand electrical signalling between neighboring cells in a biofilm.
For more information about the mathematical theory of percolation, Kim Christensen’s notes on the subject provide a detailed introduction.
- Bak, P., Tang, C., & Wiesenfeld, K. (1987). Self-organized criticality: An explanation of the 1/fnoise. Physical Review Letters, 59(4), 381–384. 10.1103/physrevlett.59.381
- Bak, P., & Tang, C. (1989). Earthquakes as a self‐organized critical phenomenon. Journal of Geophysical Research: Solid Earth, 94(B11), 15635–15637. 10.1029/jb094ib11p15635
- Hesse, J., & Gross, T. (2014). Self-organized criticality as a fundamental property of neural systems. Frontiers in Systems Neuroscience, 8. 10.3389/fnsys.2014.00166
- Pla, O., & Nori, F. (1991). Self-Organized Critical Behavior in Pinned Flux Lattices. Physical Review Letters, 67(7), 919–922. 10.1103/physrevlett.67.919
- Kramer, S., & Marder, M. (1992). Evolution of river networks. Physical Review Letters, 68(2), 205–208. 10.1103/physrevlett.68.205
- Pietronero, L., Vespignani, A., & Zapperi, S. (1994). Renormalization scheme for self-organized criticality in sandpile models. Physical Review Letters, 72(11), 1690–1693. 10.1103/physrevlett.72.1690
- Redner, S., & Mueller, P. R. (1982). Conductivity of a random directed-diode network near the percolation threshold. Physical Review B, 26(9), 5293–5295. 10.1103/physrevb.26.5293
- Takeuchi, K. A., Kuroda, M., Chaté, H., & Sano, M. (2007). Directed Percolation Criticality in Turbulent Liquid Crystals. Physical Review Letters, 99(23). 10.1103/physrevlett.99.234503
- Zhai, X., Larkin, J. W., Kikuchi, K., Redford, S. E., Roy, U., Süel, G. M., & Mugler, A. (2019). Statistics of correlated percolation in a bacterial community. PLOS Computational Biology, 15(12), e1007508. 10.1371/journal.pcbi.1007508
