Deep neural networks and decision boundaries¶
Preamble: Run the cells below to import the necessary Python packages
This notebook created by William Gilpin. Consult the course website for all content and GitHub repository for raw files and runnable online code.
## Preamble / required packages
import numpy as np
np.random.seed(0)
## Import local plotting functions and in-notebook displazooy functions
import matplotlib.pyplot as plt
from IPython.display import Image, display
%matplotlib inline
import warnings
## Comment this out to activate warnings
warnings.filterwarnings('ignore')
Is there anything linear models can't do?¶
We previously saw that regularized linear models can be surprisingly effective for supervised learning tasks like forecasting, regression, and even classification.
In unsupervised learning, PCA and its variants can be thought of as linear models
Are there any problems that are fundamentally beyond the capabilities of linear models?
## AND dataset for training
X = np.array([[0.0, 0], [0, 1], [1, 0], [1, 1]])
y = np.array([0, 0, 0.0, 1.0])
print("Training data has shape: ", X.shape)
print("Target targets have shape: ", y.shape)
## Plot the AND dataset
plt.figure(figsize=(5, 5))
plt.scatter(X[:, 0], X[:, 1], c=y, cmap=plt.cm.coolwarm, s=500)
plt.xlabel("X Feature 1")
plt.ylabel("X Feature 2")
plt.colorbar(label="Target")
Training data has shape: (4, 2) Target targets have shape: (4,)
<matplotlib.colorbar.Colorbar at 0x1179ac2d0>
Since there are discrete-valued labels for each data point, we can think of this as a classification problem.
We will now fit a logistic regression model to this data, and then visualize the predicted test labels
## train logistic regression model
from sklearn.linear_model import LogisticRegression
model = LogisticRegression(penalty='none')
model.fit(X, y)
yhat = model.predict(X)
print("Predicted labels have shape: ", yhat.shape)
print("Train accuracy: ", np.mean(yhat == y))
plt.figure(figsize=(5, 5))
plt.scatter(X[:, 0], X[:, 1], c=yhat, cmap=plt.cm.coolwarm, s=500)
plt.xlabel("Feature 1")
plt.ylabel("Feature 2")
print(yhat)
Predicted labels have shape: (4,) Train accuracy: 1.0 [0. 0. 0. 1.]
Decision boundaries¶
In the above plot, we visualized the predicted labels specifically for the training datapoints
However, in principle the training features can have any values in the feature space, and so we can construct a testing dataset spanning the domain of the feature space.
For a trained classification model, labelling each point in this domain reveals the decision boundary of the model
## plot decision boundary
def plot_decision_boundary(X, y, clf):
"""
Plot the decision boundary of a trained classifier clf
Args:
X (numpy.ndarray): Input data
y (numpy.ndarray): Input labels
clf (sklearn.base.BaseEstimator): Trained classifier
Returns:
None
"""
# Set min and max values and give it some padding
x_min, x_max = X[:, 0].min() - .5, X[:, 0].max() + .5
y_min, y_max = X[:, 1].min() - .5, X[:, 1].max() + .5
h = 0.01
# Generate a grid of points with distance h between them
xx, yy = np.meshgrid(np.arange(x_min, x_max, h), np.arange(y_min, y_max, h))
# Predict the function value for the whole gid
Z = clf.predict(np.c_[xx.ravel(), yy.ravel()])
# Put the result into a color plot
Z = Z.reshape(xx.shape)
# Plot the contour and training examples
plt.contourf(xx, yy, Z, cmap='bwr')
plt.scatter(X[:, 0], X[:, 1], c=y, cmap='bwr')
plt.figure(figsize=(5, 5))
plot_decision_boundary(X, y, model)
plt.xlabel("Feature 1")
plt.ylabel("Feature 2")
plt.title("Decision boundary for Logistic classifier on AND dataset")
Text(0.5, 1.0, 'Decision boundary for Logistic classifier on AND dataset')
Questions¶
Why is the decision boundary a straight line?
What might influence the location and slope of the decision boundary?
Another simple dataset¶
## XOR dataset for training
X = np.array([[0, 0], [0, 1], [1, 0], [1, 1]])
y = np.array([0, 1.0, 1.0, 0.0])
## Plot the XOR dataset
plt.figure(figsize=(5, 5))
plt.scatter(X[:, 0], X[:, 1], c=y, cmap=plt.cm.coolwarm, s=500)
plt.xlabel("Feature 1")
plt.ylabel("Feature 2")
print(X.shape)
print(y.shape)
(4, 2) (4,)
## train logistic regression model
from sklearn.linear_model import LogisticRegression
## XOR dataset for training
X = np.array([[0, 0], [0, 1], [1, 0], [1, 1]])
y = np.array([0, 1.0, 1.0, 0.0])
model = LogisticRegression(penalty='none')
model.fit(X, y)
yhat = model.predict(X)
print("Predicted labels have shape: ", yhat.shape)
print("Train accuracy: ", np.mean(yhat == y))
Predicted labels have shape: (4,) Train accuracy: 0.5
# plt.figure()
# plt.scatter(X[:, 0], X[:, 1], c=yhat, cmap=plt.cm.coolwarm, s=500)
# plt.xlabel("Feature 1")
# plt.ylabel("Feature 2")
plt.figure(figsize=(5, 5))
plot_decision_boundary(X, y, model)
plt.xlabel("Feature 1")
plt.ylabel("Feature 2")
plt.title("Decision boundary for Logistic classifier on XOR dataset")
Text(0.5, 1.0, 'Decision boundary for Logistic classifier on XOR dataset')
What is a neural network?¶
- A neural network is a function that takes a vector of inputs and returns a vector of outputs
- Anything for which you can use a matrix, you can use a neural network
- Like linear regression, the entries in the matrix are trainable parameters
Linear regression¶
$$ \hat{y} = \boldsymbol{\theta} X $$
Generalized linear regression¶
$$ \hat{y} = \sigma(\boldsymbol{\theta} X) $$ where $\sigma(\cdot)$ is a nonlinear "link" function. For logistic function, $\sigma(u) = {1}/({1 + e^{-u}})$
Multilayer perceptron (a neural network)¶
$$ \hat{y} = \sigma(\boldsymbol{\theta}_2 \sigma(\boldsymbol{\theta}_1 X)) $$
where $\sigma$ is a nonlinear function, and $\boldsymbol{\theta}$ is a matrix of trainable parameters.
## TODO: Add noise to inputs
from sklearn.neural_network import MLPClassifier
model = MLPClassifier(hidden_layer_sizes=(5, 300, 300, 5), activation='tanh', max_iter=1000, random_state=0)
## XOR dataset for training
X = np.array([[0, 0], [0, 1], [1, 0], [1, 1]])
y = np.array([0, 1.0, 1.0, 0.0])
## AND
# X = np.array([[0, 0], [0, 1], [1, 0], [1, 1]])
# y = np.array([0, 0.0, 0.0, 1.0])
model.fit(X, y)
yhat = model.predict(X)
plt.figure(figsize=(5, 5))
plt.scatter(X[:, 0], X[:, 1], c=yhat, cmap=plt.cm.coolwarm, s=500)
plt.xlabel("Feature 1")
plt.ylabel("Feature 2")
plt.figure(figsize=(5, 5))
plot_decision_boundary(X, yhat, model)
plt.xlabel("Feature 1")
plt.ylabel("Feature 2")
Text(0, 0.5, 'Feature 2')
## Read out the weight matrices
# model.coefs_
for layer in model.coefs_:
print(layer.shape)
(2, 5) (5, 5) (5, 1)
What do neural networks do?¶
- Neural networks can learn "arbitrary" nonlinear functions of the data
- Cybenko's theorem: any continuous function can be approximated by a neural network with a single, sufficiently wide hidden layer
- Modern generalizations exist for deep networks, etc
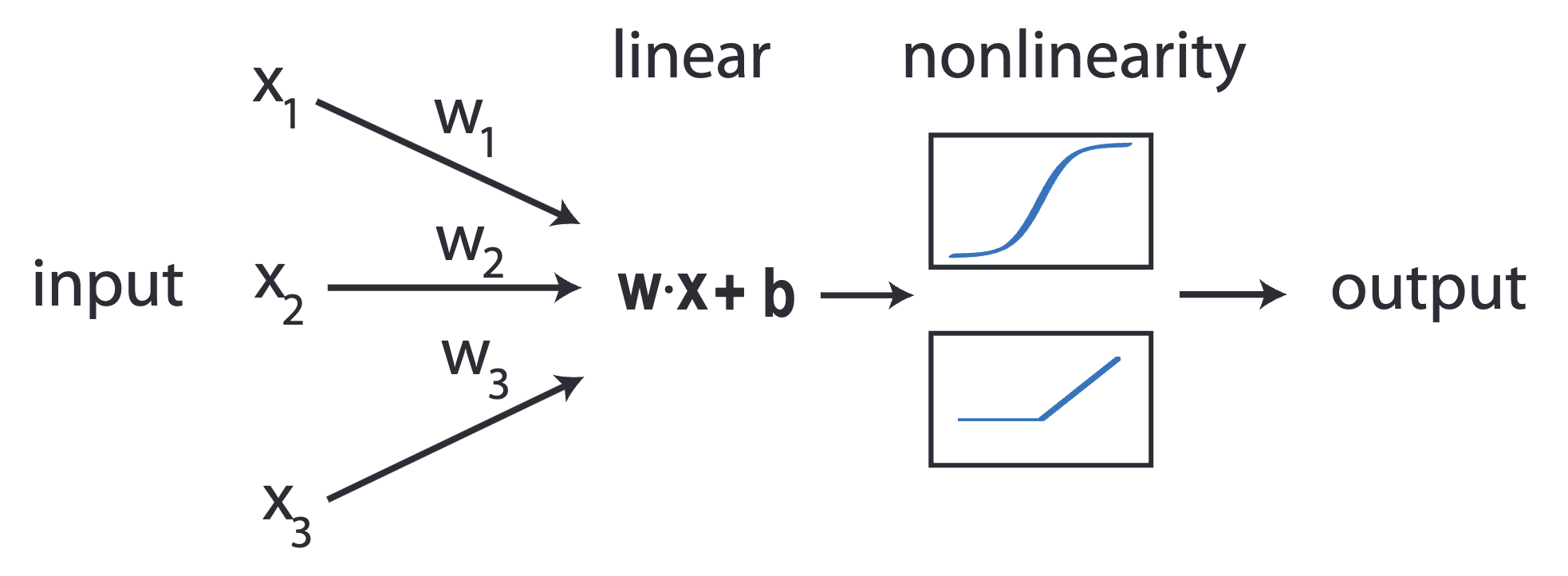
Multilayer perceptron¶
Simplest neural network: multilayer perceptron: matrix multiplication followed by a nonlinear function
Hyperparameters: depth is number of layers, width is number of "neurons" (units) per layer, activation function, regularization, etc
The number of trainable parameters is the number of entries across all matrices. Overparameterized in many cases.
Image('../resources/mlp.png')
# https://medium.com/codex/introduction-to-how-an-multilayer-perceptron-works-but-without-complicated-math-a423979897ac
The diagram above is equivalent to writing
$$ \hat{\mathbf{y}} = \sigma(\boldsymbol{\theta}_3\sigma(\boldsymbol{\theta}_2 \sigma(\boldsymbol{\theta}_1 X))) $$
where $\boldsymbol{\theta}_1 \in \mathbb{R}^{5 \times 5}$, $\boldsymbol{\theta}_2 \in \mathbb{R}^{5 \times 5}$, $\boldsymbol{\theta}_3 \in \mathbb{R}^{5 \times 2}$
## The code version
def mlp_forward(X):
theta1 = np.random.random((5, 5))
theta2 = np.random.random((5, 5))
theta3 = np.random.random((5, 2))
h1 = np.tanh(X @ theta1)
h2 = np.tanh(h1 @ theta2)
h3 = np.tanh(h2 @ theta3)
return h3
X = np.random.random((10000, 5))
print("Input shape: ", X.shape)
print("Output shape: ", mlp_forward(X).shape)
Input shape: (10000, 5) Output shape: (10000, 2)
The choice of activation function is itself a hyperparameter¶
import numpy as np
import matplotlib.pyplot as plt
# Define the activation functions
def hopfield(z):
return np.where(z >= 0, 1, 0)
def sigmoid(z):
return 1 / (1 + np.exp(-z))
def tanh(z):
return np.tanh(z)
def relu(z):
return np.maximum(0, z)
def leaky_relu(z, alpha=0.1):
return np.where(z > 0, z, alpha * z)
def elu(z, alpha=1.0):
return np.where(z > 0, z, alpha * (np.exp(z) - 1))
def selu(z, lambda_=1.0507, alpha=1.67326):
return np.where(z > 0, lambda_ * z, lambda_ * alpha * (np.exp(z) - 1))
def silu(z):
return z / (1 + np.exp(-z))
def softplus(z):
return np.log(1 + np.exp(z))
# Set up the figure and axis
z = np.linspace(-5, 5, 100)
fig, axs = plt.subplots(3, 3, figsize=(12, 9), facecolor='black')
# fig.subplots_adjust(hspace=0.5, wspace=0.4)
fig.subplots_adjust(hspace=0.6, wspace=0.4, top=0.92, bottom=0.08)
# Define titles and functions for each subplot
titles = [
'Hopfield', 'Sigmoid', 'Tanh', 'ReLU', 'Leaky ReLU', 'ELU',
'SELU', 'SiLU', 'Softplus'
]
functions = [
hopfield, sigmoid, tanh, relu, leaky_relu, elu,
selu, silu, softplus
]
equations = [
r"$\Theta(z)$",
r"$\frac{1}{1+e^{-z}}$",
r"$\tanh(z)$",
r"$\max(0, z)$",
r"$0.1z$ if $z \leq 0$, $z$ if $z > 0$",
r"$e^z - 1$ if $z \leq 0$, $z$ if $z > 0$",
r"$\lambda z$ if $z > 0$, $\lambda \alpha (e^z - 1)$ if $z \leq 0$",
r"$z \cdot \sigma(z)$ where $\sigma(z) = \frac{1}{1 + e^{-z}}$",
r"$\log(1 + e^z)$"
]
# Plot each activation function
for i, (ax, title, func, equation) in enumerate(zip(axs.flat, titles, functions, equations)):
ax.plot(z, func(z), color='orange', linewidth=2)
ax.set_title(title, fontsize=16, color='white')
ax.text(0, -0.5, equation, fontsize=12, color='white', ha='center')
ax.grid(False)
ax.set_facecolor('black')
ax.spines['bottom'].set_color('white')
ax.spines['left'].set_color('white')
ax.tick_params(axis='x', colors='white')
ax.tick_params(axis='y', colors='white')
if i < 3:
ax.set_ylim(-1, 1.1)
elif i < 6:
ax.set_ylim(-1, 6)
else:
ax.set_ylim(-1.8, 6)
plt.show()
A harder classification problem: predicting the Reynolds number of turbulent flows¶
We will use a dataset of videos of 2D turbulent flows simulated at different Reynolds numbers
This dataset has spatial noise, which is a common artifact of the PIV algorithm used to extract a velocity field from videos of a flow
Each frame we will treat as a single datapoint in our dataset, with a number of features equal to the number of pixels in the frame
We will focus on the far-field wake of a cylinder, rather than the near-field, since this is a harder problem that is less obvious to the naked eye
## load the turbulence dataset
all_vorticity_fields = list()
all_reynolds_numbers = list()
# Load simulations for different Reynolds numbers
re_vals = [300, 600, 900, 1200]
for re_val in re_vals:
# Load the two-dimensional velocity field data. Data is stored in a 4D numpy array,
# where the first dimension is the time index, the second and third dimensions are the
# x and y coordinates, and the fourth dimension is the velocity components (ux or uv).
vfield = np.load(
f"../resources/von_karman_street/vortex_street_velocities_Re_{re_val}_largefile.npz",
allow_pickle=True
)
# Calculate the vorticity, which is the curl of the velocity field
vort_field = np.diff(vfield, axis=1)[..., :-1, 1] + np.diff(vfield, axis=2)[:, :-1, :, 0]
# Downsample the dataset
vort_field = vort_field[::6, -127:, :]
# Add random experimental noise
noise_field = np.random.normal(0, 0.02, vort_field.shape)
## Gaussian blur the noise field along the x and y axes using scipy.ndimage.gaussian_filter
from scipy.ndimage import gaussian_filter
noise_field = gaussian_filter(noise_field, sigma=(0, 4, 4))
vort_field += noise_field
all_vorticity_fields.append(vort_field)
all_reynolds_numbers.extend(re_val * np.ones(vort_field.shape[0]))
all_vorticity_fields = np.vstack(all_vorticity_fields)
all_reynolds_numbers = np.array(all_reynolds_numbers)
print("Vorticity field data has shape: {}".format(all_vorticity_fields.shape))
print("Reynolds number data has shape: {}".format(all_reynolds_numbers.shape))
## Plot 4x4 grid of vorticity fields
fig, ax = plt.subplots(4, 4, figsize=(8, 8))
for i in range(4):
re_val = re_vals[i]
for j in range(4):
ax[j, i].imshow(
all_vorticity_fields[all_reynolds_numbers == re_val][150 * j],
cmap='bwr',
vmin=-0.01, vmax=0.01
)
ax[j, i].axis('off')
if j == 0:
ax[j, i].set_title(f"Re = {re_val}")
plt.tight_layout()
fig.subplots_adjust(wspace=0.1, hspace=0.1)
Vorticity field data has shape: (2000, 127, 127) Reynolds number data has shape: (2000,)
Flattening¶
Our images have resolution 127x127, so we have 16129 features per frame. We will use a dataset of 1000 frames, and we will need to flatten each frame into a vector of length 16129.
## Convert into a machine-learning dataset by flattening features
# Flatten the vorticity field data
X = np.reshape(all_vorticity_fields, (all_vorticity_fields.shape[0], -1))
# Standardize the data
X = (X - np.mean(X, axis=0)) / np.std(X, axis=0)
y = all_reynolds_numbers
print("Training data has shape: {}".format(X.shape))
print("Training labels have shape: {}".format(y.shape))
Training data has shape: (2000, 16129) Training labels have shape: (2000,)
How hard is this problem? Try playing with unsupervised embeddings to evaluate the difficulty¶
Before we train a supervised model, let's try to visualize the data using some unsupervised learning techniques
Unsupervised techniques can tell us how hard the problem is, and what kind of structure the data has
Question: Why are these lines?
from sklearn.decomposition import PCA
pca = PCA(n_components=2)
X_pca = pca.fit_transform(X)
plt.figure(figsize=(9, 8))
plt.scatter(X_pca[:, 0], X_pca[:, 1], c=all_reynolds_numbers)
plt.xlabel("PCA component 1")
plt.ylabel("PCA component 2")
plt.colorbar(label="Reynolds number")
<matplotlib.colorbar.Colorbar at 0x339e0c950>
We can also try to visualize the data using a nonlinear embedding technique like UMAP. This method generally does not preserve distances (isometry), but the nearest neighbors of a point in the full 16,000 dimensional space will be preserved in the 2D embedding.
import umap
# Reduce the dimensionality of the data using UMAP
reducer = umap.UMAP(random_state=0)
X_umap = reducer.fit_transform(X)
# Plot the UMAP embedding
plt.figure(figsize=(8, 8))
plt.scatter(X_umap[:, 0], X_umap[:, 1], c=all_reynolds_numbers)
plt.xlabel("UMAP component 1")
plt.ylabel("UMAP component 2")
Text(0, 0.5, 'UMAP component 2')
Can we use our domain knowledge to better separate the data?¶
Inductive biases are when we use our problem knowledge to reduce the number of possible learning models
Like the bias-variance tradeoff, inductive biases allow us to use domain knowledge to "guide" a model, at the expense of flexibility for different problems
For the fluid flow problem, we know that the Navier-Stokes equations contain terms that are quadratic in the velocity field, as well as gradients of the velocity field
We will first try using finite time differences to featurize the data, followed by spatial fourier transforms, which implicitly give information about spatial gradients
# Try finite differences
fd1 = np.gradient(all_vorticity_fields, axis=1).reshape((all_vorticity_fields.shape[0], -1))
fd2 = np.gradient(all_vorticity_fields, axis=2).reshape((all_vorticity_fields.shape[0], -1))
X_fd = np.hstack((fd1, fd2))
X_fd = np.reshape(X_fd, (X_fd.shape[0], -1))
## Augment the feature space with finite differences
X_aug = np.concatenate([X, X_fd], axis=1)
# Try PCA and UMAP on the Fourier coefficients
# Reduce the dimensionality of the data using UMAP
reducer = PCA(n_components=2)
X_pca = reducer.fit_transform(X_fd)
plt.figure(figsize=(8, 8))
plt.scatter(X_pca[:, 0], X_pca[:, 1], c=all_reynolds_numbers)
plt.xlabel("PCA 1")
plt.ylabel("PCA 2")
reducer = umap.UMAP(random_state=0)
X_umap = reducer.fit_transform(X_fd)
plt.figure(figsize=(8, 8))
plt.scatter(X_umap[:, 0], X_umap[:, 1], c=all_reynolds_numbers)
plt.xlabel("UMAP 1")
plt.ylabel("UMAP 2")
Text(0, 0.5, 'UMAP 2')
## Try featurizing with 2D Fourier coefficients
# Calculate the 2D Fourier coefficients
X_fft = np.fft.fft2(all_vorticity_fields)
# Convert to power spectrum
X_fft = np.reshape(np.abs(X_fft)**2, (X_fft.shape[0], -1))
# Try PCA and UMAP on the Fourier coefficients
# Reduce the dimensionality of the data using UMAP
reducer = PCA(n_components=2)
X_pca = reducer.fit_transform(X_fft)
plt.figure(figsize=(8, 8))
plt.scatter(X_pca[:, 0], X_pca[:, 1], c=all_reynolds_numbers)
reducer = umap.UMAP(random_state=0)
X_umap = reducer.fit_transform(X_fft)
plt.figure(figsize=(8, 8))
plt.scatter(X_umap[:, 0], X_umap[:, 1], c=all_reynolds_numbers)
<matplotlib.collections.PathCollection at 0x3e0c8c210>
Let's try training a model to predict Reynolds number¶
As a baseline, we'll use multinomial logistic regression, which extends logistic regression to more than two classes
Generally, we prefer using a simple model as a baseline, before moving to more complex models
In the machine learning literature, baselines and ablations are important for establishing the value of a new model or architecture
from sklearn.linear_model import LogisticRegression
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
## Shuffle the data and split into training and test sets
sel_inds = np.random.permutation(X.shape[0])[:400]
X_all, y_all = X[sel_inds], y[sel_inds]
X_train, X_test, y_train, y_test = train_test_split(X_all, y_all, test_size=0.4, random_state=0)
# X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.4, random_state=0)
model_logistic = LogisticRegression()
model_logistic.fit(X_train, y_train)
y_pred_logistic = model_logistic.predict(X_test)
print("Training set score: {:.2f}".format(model_logistic.score(X_train, y_train)))
print("Test set score: {:.2f}".format(model_logistic.score(X_test, y_test)))
plt.imshow(confusion_matrix(y_test, y_pred_logistic))
plt.xlabel("Predicted")
plt.ylabel("True")
plt.xticks(np.arange(len(re_vals)), re_vals);
plt.yticks(np.arange(len(re_vals)), re_vals);
Training set score: 1.00 Test set score: 0.62
from sklearn.neural_network import MLPClassifier
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.4, random_state=0)
# A 4 layer neural network with 10 hidden units in each layer
mlp = MLPClassifier(hidden_layer_sizes=(3, 3), random_state=0)
mlp.fit(X_train, y_train)
y_pred = mlp.predict(X_test)
print("Training set score: {:.2f}".format(mlp.score(X_train, y_train)))
print("Test set score: {:.2f}".format(mlp.score(X_test, y_test)))
plt.imshow(confusion_matrix(y_test, y_pred))
plt.xlabel("Predicted")
plt.ylabel("True")
plt.xticks(np.arange(len(re_vals)), re_vals);
plt.yticks(np.arange(len(re_vals)), re_vals);
Training set score: 0.89 Test set score: 0.76
Can we do better? How deep should we go?¶
Model choices = hyperparameter tuning. Recall that any "choice" that affects model complexity is a hyperparameter, including the choice of model itself, the optimizer, the learning rate, the number of layers, the number of neurons per layer, the activation function, etc. Here, we will focus on the number and width of layers
We use cross-validation (we split train into validation sets)
To search over hyperparameters, we use scikit-learn's
GridSearchCV, which uses the same API as a standard model, but which internally trains and evaluates the model on all possible hyperparameter combinations with cross-validation
## Tuning hyperparameters with sklearn built-in grid search and cross-validation
from sklearn.model_selection import GridSearchCV
param_grid = {'hidden_layer_sizes': [(3, 3, 3), (5, 5), (7, 5, 3)]}
grid = GridSearchCV(MLPClassifier(), param_grid, cv=5)
grid.fit(X_train, y_train)
print("Best cross-validation score: {:.2f}".format(grid.best_score_))
print("Best parameters: ", grid.best_params_)
print("Test set score: {:.2f}".format(grid.score(X_test, y_test)))
plt.imshow(confusion_matrix(y_test, grid.predict(X_test)))
plt.xlabel("Predicted")
plt.ylabel("True")
plt.xticks(np.arange(len(re_vals)), re_vals);
plt.yticks(np.arange(len(re_vals)), re_vals);
Best cross-validation score: 0.79
Best parameters: {'hidden_layer_sizes': (5, 5)}
Test set score: 0.83
## Use the best hyperparameters
mlp = MLPClassifier(**grid.best_params_, random_state=0)
mlp.fit(X_train, y_train)
y_pred_train = mlp.predict(X_train)
y_pred_test = mlp.predict(X_test)
print("Training set score: {:.2f}".format(mlp.score(X_train, y_train)))
print("Test set score: {:.2f}".format(mlp.score(X_test, y_test)))
plt.imshow(confusion_matrix(y_test, y_pred))
plt.xlabel("Predicted")
plt.ylabel("True")
plt.xticks(np.arange(len(re_vals)), re_vals);
plt.yticks(np.arange(len(re_vals)), re_vals);
Training set score: 1.00 Test set score: 0.82
Visualize the decision boundary¶
Our images live in a high-dimensional feature space, but we can still visualize the decision boundary in a lower-dimensional space using the embedding techniques we learned about last week
We will visualize our decision boundary in this embedding space
# Train embedding on the data
reducer = umap.UMAP(random_state=0)
reducer.fit(X)
X_umap_train = reducer.transform(X_train)
X_umap_test = reducer.transform(X_test)
plt.figure(figsize=(8, 8))
plt.scatter(X_umap_train[:, 0], X_umap_train[:, 1], c=y_train)
plt.scatter(X_umap_test[:, 0], X_umap_test[:, 1], c=y_test, marker='o')
plt.xlabel("UMAP 1")
plt.ylabel("UMAP 2")
plt.title("Ground Truth")
plt.figure(figsize=(8, 8))
plt.scatter(X_umap_train[:, 0], X_umap_train[:, 1], c=y_pred_train)
plt.scatter(X_umap_test[:, 0], X_umap_test[:, 1], c=y_pred_test, marker='o')
plt.xlabel("UMAP 1")
plt.ylabel("UMAP 2")
plt.title("Multi-Layer Perceptron")
plt.figure(figsize=(8, 8))
# plt.scatter(X_umap_train[:, 0], X_umap_train[:, 1], c=y_train)
# plt.scatter(X_umap_test[:, 0], X_umap_test[:, 1], c=y_pred_logistic, marker='o')
# plt.xlabel("UMAP 1")
# plt.ylabel("UMAP 2")
# plt.title("Logistic Regression")
<Figure size 800x800 with 0 Axes>
<Figure size 800x800 with 0 Axes>
How big is our model?¶
Our final neural network maps from $127^2 = 16,129$ features to 4 classes, with 1 hidden layers containing 10 neurons
This means we have around 160,000 trainable parameters
grid.best_params_
{'hidden_layer_sizes': (7,)}
A convolutional neural network¶
Can we process the images in a more parameter-efficient way?
The Multi-Layer oerceptron model we used above has a lot of trainable parameters, since it maps flattened images that have $127^2 = 16129$ features into a single output. Even a linear model would have 16129 parameters!
We know that turbulent flows have a lot of spatial structure, and that the Navier-Stokes equations are local in space. Can we build this property into our model?
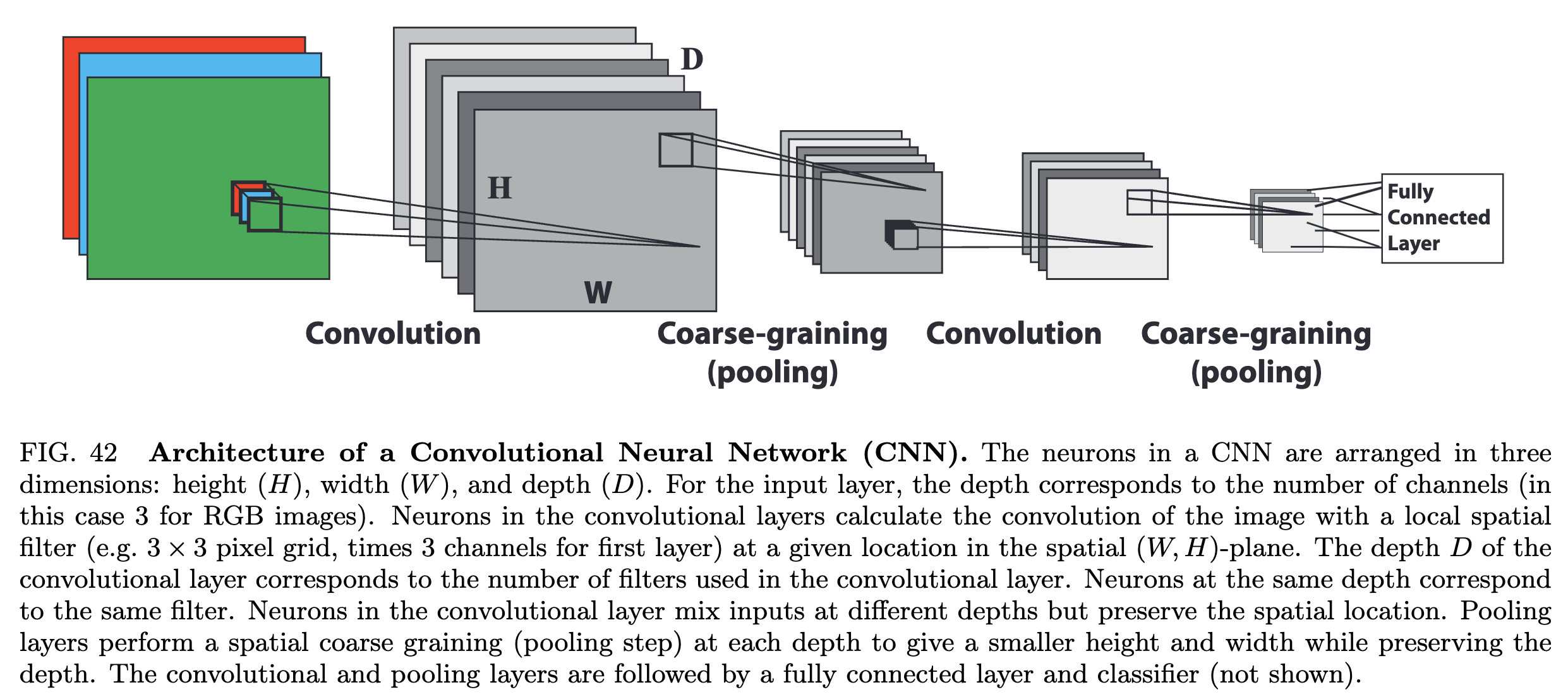
Image from Mehta et al. 2018
In a convolutional neural network (CNN), we avoid explicitly flattening the image, and instead apply a series of trainable convolutional filters to the image
Recall that convolutional filters are usually small kernels, like $3 \times 3$ or $5 \times 5$ images, that we slide across the image. The output of a discrete convolution is a new image, where each pixel is a combination of pixel neighborhoods from the previous image
Our image classifier needs to map from an image to a single output, and so CNN also includes a series of pooling layers, which downsample the intermediate images
After the image becomes sufficiently small, we then flatten it and apply a standard fully-connected neural network to the flattened image
- As a starting point, we will leave our input dataset in the form of images. So instead of our training data having the shape $X \in \mathbb{R}^{N_\text{data} \times N_\text{feats}}$, it will have the shape $X \in \mathbb{R}^{N_\text{data} \times N_\text{pixels} \times N_\text{pixels}}$
## Convert into a machine-learning dataset by flattening features
X = np.copy(all_vorticity_fields)[..., None]
# Standardize the data
X = (X - np.mean(X, axis=0)) / np.std(X, axis=0)
y = np.unique(all_reynolds_numbers, return_inverse=True)[1] # Convert labels to integers
from sklearn.model_selection import train_test_split
from sklearn.metrics import confusion_matrix
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.4, random_state=0)
print("Training data has shape: {}".format(X_train.shape))
print("Training labels have shape: {}".format(y_train.shape))
print("Test data has shape: {}".format(X_test.shape))
print("Test labels have shape: {}".format(y_test.shape))
plt.hist(y_train)
plt.xlabel("Class")
plt.ylabel("Count")
Training data has shape: (1200, 127, 127, 1) Training labels have shape: (1200,) Test data has shape: (800, 127, 127, 1) Test labels have shape: (800,)
Text(0, 0.5, 'Count')
We will now implement a CNN using JAX, a library for automatic differentiation and GPU acceleration
Our implementation will inherit structure from scikit-learn, so that we can use the same fit and predict API as before
import jax
import jax.numpy as jnp
from jax import grad, jit, random, value_and_grad
import optax # An optimization library for JAX
from sklearn.base import BaseEstimator, ClassifierMixin
class CNNClassifier(BaseEstimator, ClassifierMixin):
"""
A Convolutional Neural Network (CNN) classifier implemented using JAX and NumPy.
Parameters:
learning_rate (float): The learning rate for the optimizer
epochs (int): The number of training epochs
batch_size (int): The batch size for training
random_state (int): The random seed for reproducibility
"""
def __init__(self, learning_rate=0.001, epochs=10, batch_size=32, random_state=0, store_history=True):
## Set hyperparameters
self.learning_rate = learning_rate
self.epochs = epochs
self.batch_size = batch_size
## Set trainable parameters and internal object state
self.params = None
self.opt_state = None
self.random_state = random_state
# JAX uses PRNG objects to control random number generation
self.rng = random.PRNGKey(random_state)
self.store_history = store_history
if self.store_history:
self.loss_history = []
def _init_params(self, input_shape, num_classes):
"""
Initialize the model parameters
Args:
input_shape (tuple): The shape of the input data
num_classes (int): The number of classes in the classification task
Returns:
None
"""
self.params = {
"conv1": {
"w": random.normal(self.rng, (3, 3, input_shape[-1], 32)),
"b": jnp.zeros((32,))
},
"fc": {
"w": random.normal(self.rng, (32 * (input_shape[0] // 2) * (input_shape[1] // 2), num_classes)),
"b": jnp.zeros((num_classes,))
}
}
def _forward(self, params, X):
"""
Forward pass of the CNN model. Given a batch of data, compute the logits
corresponding to each class.
Args:
params (dict): Dictionary containing the model parameters
X (numpy.ndarray): Batch of input data
Returns:
numpy.ndarray: Logits for each class
"""
def relu(x):
return jnp.maximum(0, x)
# Convolution layer transforms the input image from a shape of
# (batch_size, height, width, channels), here (64, 128, 128, 1), to a shape of
# (batch_size, height, width, num_filters), here (64, 128, 128, 32)
conv1_out = jax.lax.conv_general_dilated(
X,
params["conv1"]["w"],
window_strides=(1, 1),
dimension_numbers=("NHWC", "HWIO", "NHWC"),
padding="SAME"
)
conv1_out = relu(conv1_out + params["conv1"]["b"])
# Pooling layer shrinks the intermediate representation to
# (batch_size, height // 2, width // 2, num_filters), here (64, 64, 64, 32)
pool_out = jax.lax.reduce_window(
conv1_out,
0.0,
jax.lax.add,
window_dimensions=(1, 2, 2, 1),
window_strides=(1, 2, 2, 1),
padding="VALID"
)
# The flattening operation reshapes the tensor to (batch_size, num_features),
# here (64, 64 * 64 * 32), and then computes the logits
flattened = pool_out.reshape((X.shape[0], -1))
logits = jnp.dot(flattened, params["fc"]["w"]) + params["fc"]["b"]
return logits
def _loss(self, params, X, y):
"""
Compute the cross-entropy loss between the model predictions and the true labels
The cross-entropy loss is defined as:
L = -1/N * sum_i sum_j y_ij * log(p_ij)
where N is the number of samples, y_ij is 1 if sample i belongs to class j, and p_ij
is the predicted probability that sample i belongs to class j.
Args:
params (dict): Dictionary containing the model parameters
X (numpy.ndarray): Batch of input data
y (numpy.ndarray): Batch of labels
Returns:
numpy.ndarray: The cross-entropy loss, averaged over the batch
"""
logits = self._forward(params, X)
y_onehot = jax.nn.one_hot(y, logits.shape[-1])
loss = -jnp.mean(jnp.sum(y_onehot * jax.nn.log_softmax(logits), axis=-1))
return loss
def _update(self, params, opt_state, X, y):
"""
Compute the gradients of the loss with respect to the model parameters and
update the model
Args:
params (dict): Dictionary containing the model parameters
opt_state (dict): Dictionary containing the optimizer state
X (numpy.ndarray): Batch of input data
y (numpy.ndarray): Batch of labels
Returns:
dict: Updated model parameters
dict: Updated optimizer state
float: The loss value
"""
loss, grads = value_and_grad(self._loss)(params, X, y)
updates, opt_state = self.optimizer.update(grads, opt_state, params)
params = optax.apply_updates(params, updates)
return params, opt_state, loss
def fit(self, X, y):
"""
Fit the CNN model to the training data
Args:
X (numpy.ndarray): Training data of shape (num_samples, height, width, channels)
y (numpy.ndarray): Training labels of shape (num_samples,)
"""
## Initialize the model parameters and optimizer at the first call to fit
if self.params is None:
input_shape = X.shape[1:]
num_classes = len(jnp.unique(y))
self._init_params(input_shape, num_classes)
self.optimizer = optax.adam(self.learning_rate)
self.opt_state = self.optimizer.init(self.params)
## Optimization loop
for epoch in range(self.epochs):
perm = random.permutation(self.rng, X.shape[0])
X, y = X[perm], y[perm]
for i in range(0, X.shape[0], self.batch_size):
X_batch = X[i:i + self.batch_size]
y_batch = y[i:i + self.batch_size]
self.params, self.opt_state, loss = self._update(self.params, self.opt_state, X_batch, y_batch)
if self.store_history:
self.loss_history.append(loss)
print(f"Epoch {epoch + 1}/{self.epochs}, Loss: {loss:.4f}")
def predict(self, X):
"""
Make predictions on new data
Args:
X (numpy.ndarray): New data of shape (num_samples, height, width, channels)
Returns:
numpy.ndarray: Predicted labels
"""
logits = self._forward(self.params, X)
return jnp.argmax(logits, axis=-1)
## Fit the CNN model
# # Train and predict
model = CNNClassifier(learning_rate=0.0001, epochs=100, batch_size=16)
model.fit(X_train, y_train)
plt.plot(model.loss_history)
plt.xlabel("Epoch")
plt.ylabel("Cross-entropy loss on training set")
Epoch 1/100, Loss: 2344.1562 Epoch 2/100, Loss: 1893.5312 Epoch 3/100, Loss: 2167.5793 Epoch 4/100, Loss: 1665.8159 Epoch 5/100, Loss: 1502.6653 Epoch 6/100, Loss: 948.9930 Epoch 7/100, Loss: 1231.2178 Epoch 8/100, Loss: 1153.6851 Epoch 9/100, Loss: 929.2589 Epoch 10/100, Loss: 652.1995 Epoch 11/100, Loss: 492.6980 Epoch 12/100, Loss: 784.1656 Epoch 13/100, Loss: 455.9197 Epoch 14/100, Loss: 227.0690 Epoch 15/100, Loss: 274.9376 Epoch 16/100, Loss: 538.9522 Epoch 17/100, Loss: 682.2462 Epoch 18/100, Loss: 94.5268 Epoch 19/100, Loss: 242.6960 Epoch 20/100, Loss: 158.2983 Epoch 21/100, Loss: 236.4017 Epoch 22/100, Loss: 401.7467 Epoch 23/100, Loss: 113.3213 Epoch 24/100, Loss: 195.1394 Epoch 25/100, Loss: 117.2153 Epoch 26/100, Loss: 113.0649 Epoch 27/100, Loss: 261.6731 Epoch 28/100, Loss: 215.6727 Epoch 29/100, Loss: 132.2099 Epoch 30/100, Loss: 216.4857 Epoch 31/100, Loss: 127.0312 Epoch 32/100, Loss: 17.0016 Epoch 33/100, Loss: 165.9627 Epoch 34/100, Loss: 63.9287 Epoch 35/100, Loss: 151.2602 Epoch 36/100, Loss: -0.0000 Epoch 37/100, Loss: -0.0000 Epoch 38/100, Loss: 271.1634 Epoch 39/100, Loss: -0.0000 Epoch 40/100, Loss: -0.0000 Epoch 41/100, Loss: -0.0000 Epoch 42/100, Loss: 44.3639 Epoch 43/100, Loss: 0.5360 Epoch 44/100, Loss: 63.3394 Epoch 45/100, Loss: -0.0000 Epoch 46/100, Loss: 4.4885 Epoch 47/100, Loss: 11.2855 Epoch 48/100, Loss: -0.0000 Epoch 49/100, Loss: -0.0000 Epoch 50/100, Loss: 46.8023 Epoch 51/100, Loss: 2.0290 Epoch 52/100, Loss: -0.0000 Epoch 53/100, Loss: 6.1905 Epoch 54/100, Loss: 4.0004 Epoch 55/100, Loss: -0.0000 Epoch 56/100, Loss: -0.0000 Epoch 57/100, Loss: -0.0000 Epoch 58/100, Loss: -0.0000 Epoch 59/100, Loss: -0.0000 Epoch 60/100, Loss: 27.7790 Epoch 61/100, Loss: -0.0000 Epoch 62/100, Loss: -0.0000 Epoch 63/100, Loss: -0.0000 Epoch 64/100, Loss: -0.0000 Epoch 65/100, Loss: -0.0000 Epoch 66/100, Loss: -0.0000 Epoch 67/100, Loss: -0.0000 Epoch 68/100, Loss: 0.0001 Epoch 69/100, Loss: -0.0000 Epoch 70/100, Loss: -0.0000 Epoch 71/100, Loss: 0.8741 Epoch 72/100, Loss: -0.0000 Epoch 73/100, Loss: -0.0000 Epoch 74/100, Loss: -0.0000 Epoch 75/100, Loss: -0.0000 Epoch 76/100, Loss: -0.0000 Epoch 77/100, Loss: -0.0000 Epoch 78/100, Loss: -0.0000 Epoch 79/100, Loss: -0.0000 Epoch 80/100, Loss: -0.0000 Epoch 81/100, Loss: -0.0000 Epoch 82/100, Loss: -0.0000 Epoch 83/100, Loss: -0.0000 Epoch 84/100, Loss: -0.0000 Epoch 85/100, Loss: -0.0000 Epoch 86/100, Loss: -0.0000 Epoch 87/100, Loss: -0.0000 Epoch 88/100, Loss: -0.0000 Epoch 89/100, Loss: -0.0000 Epoch 90/100, Loss: -0.0000 Epoch 91/100, Loss: -0.0000 Epoch 92/100, Loss: -0.0000 Epoch 93/100, Loss: -0.0000 Epoch 94/100, Loss: -0.0000 Epoch 95/100, Loss: -0.0000 Epoch 96/100, Loss: -0.0000 Epoch 97/100, Loss: -0.0000 Epoch 98/100, Loss: -0.0000 Epoch 99/100, Loss: -0.0000 Epoch 100/100, Loss: -0.0000
Text(0, 0.5, 'Cross-entropy loss on training set')
y_pred_train = model.predict(X_train)
y_pred_test = model.predict(X_test)
print("Training set score: {:.2f}".format(np.mean(y_pred_train == y_train)))
print("Test set score: {:.2f}".format(np.mean(y_pred_test == y_test)))
plt.imshow(confusion_matrix(y_test, y_pred_test))
plt.xlabel("Predicted")
plt.ylabel("True")
plt.xticks(np.arange(len(re_vals)), re_vals);
plt.yticks(np.arange(len(re_vals)), re_vals);
Training set score: 1.00 Test set score: 0.78
How many parameters did that take?¶
The convolutional layer has $N_\text{filters} \times N_\text{pixels} \times N_\text{pixels} \times N_\text{channels}$ parameters. In our case, $N_\text{filters} = 16$, $N_\text{pixels} = 127$, $N_\text{channels} = 1$
The fully connected layer has $N_\text{filters} \times N_\text{pixels} \times N_\text{pixels} \times N_\text{channels} \times N_\text{neurons}$ parameters. In our case, $N_\text{neurons} = 2$
The total number of parameters is therefore $16 \times 127 \times 127 + 16 \times 2 = 325,858$
model.params['conv1']['w'].shape
(3, 3, 1, 32)
model.params['fc']['w'].shape
(127008, 4)
Inductive biases in machine learning¶
By using a CNN, we managed to get both higher accuracy and lower number of parameters
This is because the CNN architecture has inductive biase for datasets with spatial translation invariance
There's no free lunch: the CNN architecture is not as flexible as the MLP, and so it may not work as well on other datasets. But because our dataset has spatial structure, we can exploit this to get better performance
We can see this as a vignette of our role as physicists using machine learning: we can use our domain knowledge to guide our model choices, and to get better performance with fewer parameters. But this comes at the expense of flexibility for other datasets.
Currently, as of 2024, the "bitter lesson" of machine learning has been that domain-specific inductive biases become unnecessary as models become larger and trained on more data. As a result, advances in bioinformatics and language have been possible with models with relatively little domain-specific knowledge.
mlp.fit(X_train, y_train)
y_pred_train = mlp.predict(X_train)
y_pred_test = mlp.predict(X_test)
print("Training set score: {:.2f}".format(mlp.score(X_train, y_train)))
print("Test set score: {:.2f}".format(mlp.score(X_test, y_test)))